
In particular, aligning NT sequences may account for interrupted ORFs. Since they are more informative, NT sequences should be able to provide equally good or even better alignments than their sole AA translation. The NT sequence is thus less conserved but more informative than its AA translation.
Because of the redundancy of genetic codes, different codons encode the same AA. A coding sequence can be considered either at the nucleotide (NT) or amino acid (AA) level.
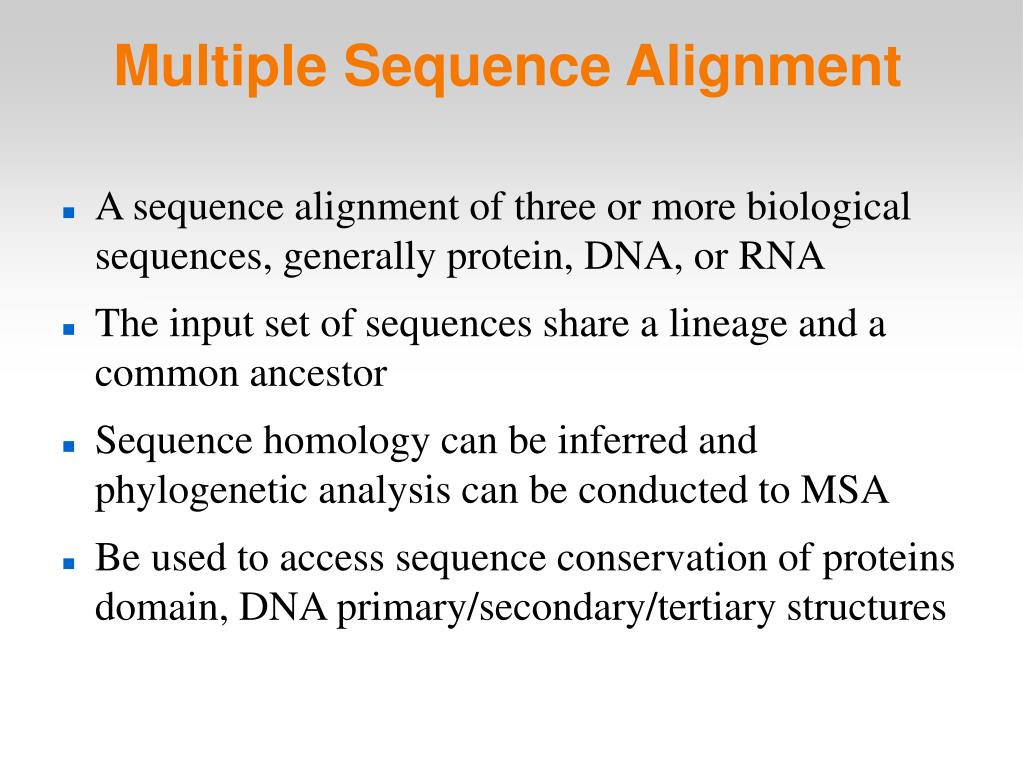
#MULTIPLE ALIGNMENT CODE#
The DNA sequences to be aligned often contain open reading frames (ORF) that code for proteins. As a consequence, MSA is a richly developed area of bioinformatics and computational biology. In all these studies, the initial MSA can strongly impact conclusions and biological interpretations. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Ĭompeting interests: The authors have declared that no competing interests exist.Ī wide range of molecular analyses rely on multiple sequence alignments (MSA), e.g., motif detection within genes and genomes, prediction of tridimensional structures, phylogenetic inference and detection of positive selection. This publication is the contribution No 2011-069 of the Institut des Sciences de l'Evolution de Montpellier (UMR 5554 - CNRS). This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.įunding: This work was supported by the French Agence Nationale de la Recherche “Domaines Emergents” (ANR-08-EMER-011, “PhylAriane”), the Centre National de la Recherche Scientifique (PEPS-INSB-2010), and the Montpellier Bioinformatics Biodiversity platform (MBB). Received: ApAccepted: JPublished: September 16, 2011Ĭopyright: © 2011 Ranwez et al. Murphy, Texas A&M University, United States of America
Ĭitation: Ranwez V, Harispe S, Delsuc F, Douzery EJP (2011) MACSE: Multiple Alignment of Coding SEquences Accounting for Frameshifts and Stop Codons. MACSE is distributed as an open-source java file executable with freely available source code and can be used via a web interface at. It has also proved useful in detecting undocumented frameshifts in public database sequences and in aligning next-generation sequencing reads/contigs against a reference coding sequence. MACSE is the first automatic solution to align protein-coding gene datasets containing non-functional sequences (pseudogenes) without disrupting the underlying codon structure. The resulting pairwise coding sequence alignment method was extended to a multiple sequence alignment (MSA) algorithm implemented in a program called MACSE (Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons). We present an algorithm that has the same space and time complexity as the classical Needleman-Wunsch algorithm while accommodating sequencing errors and other biological deviations from the coding frame. Secondly, each sequence is translated with the same reading frame from beginning to end, so that the presence of a single additional nucleotide leads to both aberrant translation and alignment.

Firstly, any premature stop codon impedes using such a strategy. There are two important pitfalls with this approach. Until now the most efficient solution to align nucleotide sequences containing open reading frames was to use indirect procedures that align amino acid translation before reporting the inferred gap positions at the codon level.


 0 kommentar(er)
0 kommentar(er)
